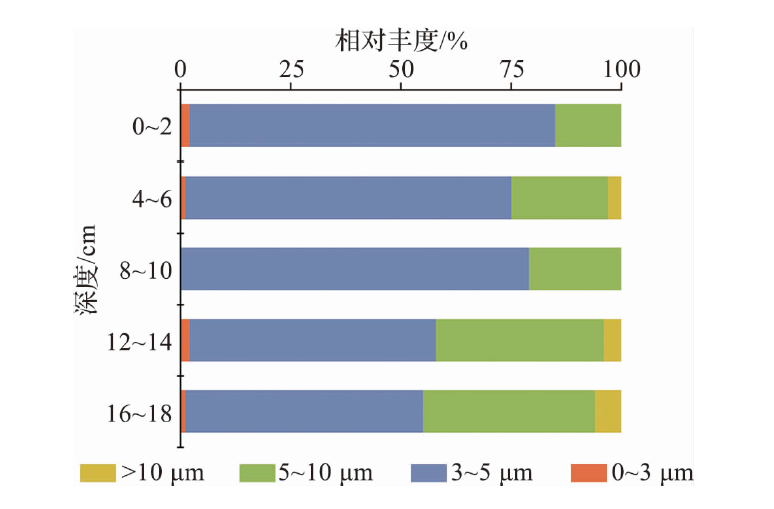
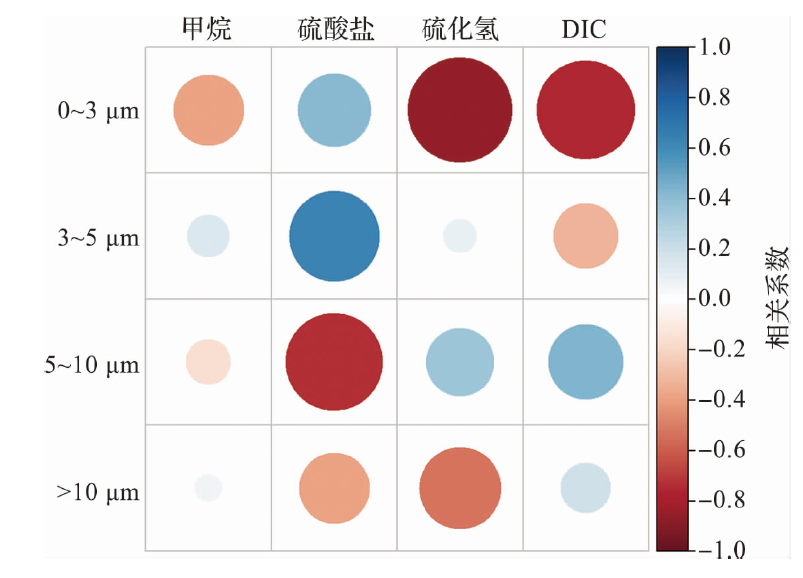
海洋学研究 ›› 2025, Vol. 43 ›› Issue (1): 22-33.DOI: 10.3969/j.issn.1001-909X.2025.01.003
应用HCR-FISH技术研究冷泉沉积物中甲烷厌氧氧化古菌的生存状态
- 上海交通大学 海洋学院,上海 200240
-
收稿日期:2024-03-22修回日期:2024-04-28出版日期:2025-03-15发布日期:2025-05-30 -
通讯作者:*梁乐文(1994—),男,博士后,主要从事海洋沉积物中微生物介导的甲烷转化过程方面的研究, E-mail: lewen94@sjtu.edu.cn。 -
作者简介:何茂雨(1999—),女,山东省日照市人,主要从事海洋沉积物环境中的微生物方面的研究,E-mail:15099986619@163.com。 -
基金资助:国家自然科学基金(42406089);中国博士后科学基金资助项目(2023M732206);国家资助博士后研究人员计划(GZB20230406)
Utilizing HCR-FISH to investigate the status of anaerobic methanotrophic archaea in cold seep sediments
HE Maoyu( ), WANG Jing, LI Sihan, LIANG Lewen*(
), WANG Jing, LI Sihan, LIANG Lewen*( )
)
- School of Oceanography, Shanghai Jiao Tong University, Shanghai 200240, China
-
Received:2024-03-22Revised:2024-04-28Online:2025-03-15Published:2025-05-30
摘要:
甲烷厌氧氧化(anaerobic oxidation of methane,AOM)过程是冷泉沉积物中元素循环过程的重要一环。该反应一般由甲烷厌氧氧化古菌(anaerobic methanotrophic archaea,ANME)和硫酸盐还原细菌(sulfate-reducing bacteria,SRB)共同完成,两者通常以共生体的方式存在。然而目前尚未获得ANME纯培养菌株,且其缓慢的代谢阻碍了对其代谢特征、协同作用机制的进一步探索和研究。本文通过杂交链反应-荧光原位杂交技术(hybridization chain reaction-fluorescence in situ hybridization,HCR-FISH)并结合16S rRNA基因高通量测序结果,调查了南海Formosa冷泉(简称F冷泉)黑色菌席区域不同深度沉积物中ANME群落组成情况及存在状态。结果显示,ANME-1和ANME-2为F冷泉沉积物中的主要ANME类群,其中,ANME-2和SRB以菌团聚集体的方式存在,而未发现ANME-1类群与细菌成团的情况。这表明ANME-2多与SRB共生进行AOM过程,并且暗示ANME-1与ANME-2存在不同的生理功能和甲烷代谢机制。另外,在所有层位的沉积物样品中,ANME-2/SRB菌团直径主要集中在3~10 μm之间。相关性分析表明,菌团直径分布与沉积物中硫酸盐浓度等环境因子显著相关,指示冷泉环境因子对ANME/SRB菌团生长的影响作用。此外,通过HCR-FISH技术进一步发现,在F冷泉沉积物中存在多个排列整齐、大小均匀的菌团连接形成的菌团簇,这种特殊结构暗示菌团间可能存在着联系或合作关系。本研究揭示了F冷泉不同深度沉积物中ANME类群的存在状态及共生菌团的大小和分布规律,为进一步揭示不同ANME类群在原位冷泉沉积物中的甲烷代谢机制和生态功能提供了基础。
中图分类号:
引用本文
何茂雨, 王景, 李思翰, 梁乐文. 应用HCR-FISH技术研究冷泉沉积物中甲烷厌氧氧化古菌的生存状态[J]. 海洋学研究, 2025, 43(1): 22-33.
HE Maoyu, WANG Jing, LI Sihan, LIANG Lewen. Utilizing HCR-FISH to investigate the status of anaerobic methanotrophic archaea in cold seep sediments[J]. Journal of Marine Sciences, 2025, 43(1): 22-33.
| 样品 编号 | 沉积物描述 | 沉积物柱状 样深度/cm | 样品数 量/个 |
|---|---|---|---|
| D231-1 | 还原性沉积物外部对照 | 14 | 7 |
| D231-2 | 还原性沉积物上边缘交界 | 30 | 15 |
| D231-3 | 还原性沉积物中心1 | 20 | 10 |
| D231-4 | 还原性沉积物中心2 | 18 | 9 |
| D231-5 | 还原性沉积物中心下边缘交界 | 22 | 11 |
| D231-6 | 还原性沉积物白色菌席 | 18 | 9 |
| 合计 | 61 | ||
表1 本研究中采用的Formosa冷泉沉积物样本信息
Tab.1 Information of sediment samples in Formosa cold seep used in this study
| 样品 编号 | 沉积物描述 | 沉积物柱状 样深度/cm | 样品数 量/个 |
|---|---|---|---|
| D231-1 | 还原性沉积物外部对照 | 14 | 7 |
| D231-2 | 还原性沉积物上边缘交界 | 30 | 15 |
| D231-3 | 还原性沉积物中心1 | 20 | 10 |
| D231-4 | 还原性沉积物中心2 | 18 | 9 |
| D231-5 | 还原性沉积物中心下边缘交界 | 22 | 11 |
| D231-6 | 还原性沉积物白色菌席 | 18 | 9 |
| 合计 | 61 | ||
| 探针名称 | 靶向类群 | 探针序列(5’-3’) | 来源 | |
|---|---|---|---|---|
| 起始 探针 | ARCH915-H | Archaea | CCGAATACAAAGCATCAACGACTAGAAAAAAGTGCTCCCCCGCCAATTCCT | 文献[ |
| EUB338-R | Bacteria | TACGCCCTAAGAATCCGAACCCTATGAAATAGCTGCCTCCCGTAGGAGT | 文献[ | |
| ANME-1-350-H | ANME-1 | CCGAATACAAAGCATCAACGACTAGAAAAAAAGTTTTCGCGCCTGATGC | 文献[ | |
| ANME-2-538-H | ANME-2 | CCGAATACAAAGCATCAACGACTAGAAAAAAGGCTACCACTCGGGCCGC | 本研究 | |
| ANME-3-1249-H | ANME-3 | CCGAATACAAAGCATCAACGACTAGAAAAAATCGGAGTAGGGACCCATT | 本研究 | |
| DSS658-R | Desulfosarcina-Desulfococcus | TACGCCCTAAGAATCCGAACCCTATGAAATATCCACTTCCCTCTCCCAT | 本研究 | |
| 扩增 探针 | H1 | CATAGGGTTCGGATTCTTAGGGCGTAGCAGCATCAATACGCCCTAAGAATCC | 文献[ | |
| H2 | TACGCCCTAAGAATCCGAACCCTATGGGATTCTTAGGGCGTATTGATGCTGC | 文献[ | ||
| R1 | TCTAGTCGTTGATGCTTTGTATTCGGCGACAGATAACCGAATACAAAGCATC | 文献[ | ||
| R2 | CCGAATACAAAGCATCAACGACTAGAGATGCTTTGTATTCGGTTATCTGTCG | 文献[ | ||
表2 本研究中使用的HCR-FISH探针序列
Tab.2 HCR-FISH probes used in this study
| 探针名称 | 靶向类群 | 探针序列(5’-3’) | 来源 | |
|---|---|---|---|---|
| 起始 探针 | ARCH915-H | Archaea | CCGAATACAAAGCATCAACGACTAGAAAAAAGTGCTCCCCCGCCAATTCCT | 文献[ |
| EUB338-R | Bacteria | TACGCCCTAAGAATCCGAACCCTATGAAATAGCTGCCTCCCGTAGGAGT | 文献[ | |
| ANME-1-350-H | ANME-1 | CCGAATACAAAGCATCAACGACTAGAAAAAAAGTTTTCGCGCCTGATGC | 文献[ | |
| ANME-2-538-H | ANME-2 | CCGAATACAAAGCATCAACGACTAGAAAAAAGGCTACCACTCGGGCCGC | 本研究 | |
| ANME-3-1249-H | ANME-3 | CCGAATACAAAGCATCAACGACTAGAAAAAATCGGAGTAGGGACCCATT | 本研究 | |
| DSS658-R | Desulfosarcina-Desulfococcus | TACGCCCTAAGAATCCGAACCCTATGAAATATCCACTTCCCTCTCCCAT | 本研究 | |
| 扩增 探针 | H1 | CATAGGGTTCGGATTCTTAGGGCGTAGCAGCATCAATACGCCCTAAGAATCC | 文献[ | |
| H2 | TACGCCCTAAGAATCCGAACCCTATGGGATTCTTAGGGCGTATTGATGCTGC | 文献[ | ||
| R1 | TCTAGTCGTTGATGCTTTGTATTCGGCGACAGATAACCGAATACAAAGCATC | 文献[ | ||
| R2 | CCGAATACAAAGCATCAACGACTAGAGATGCTTTGTATTCGGTTATCTGTCG | 文献[ | ||
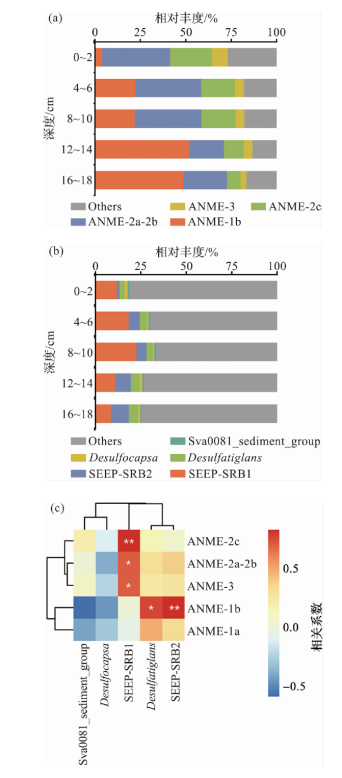
图1 Formosa冷泉沉积物样本中ANME和SRB类群的组成及相关性 (图a表示D231-4站位中ANME类群随深度的变化,图中的“Others”表示除ANME以外的古菌和ASV小于1%的ANME;图b表示D231-4站位中SRB类群随深度的变化,图中的“Others”表示除SRB以外的细菌和ASV小于1%的SRB;图c表示D231站位所有层位样本中ANME和SRB类群的Pearson相关性热图,共使用61个沉积物样品,*表示p<0.05,**表示p<0.01。)
Fig.1 Composition and correlation of ANME and SRB groups in the sediment samples of Formosa cold seep (Figure a illustrates the variation of ANME groups with depth in site D231-4, where “Others” in the figure represents archaea other than ANME and ANME with ASV abundances less than 1%. Figure b depicts the variation of SRB groups with depth in site D231-4, where “Others” denotes bacteria other than SRB and SRB with ASV abundances less than 1%. Figure c presents a Pearson correlation heatmap of ANME and SRB groups across all depth intervals from station D231, based on 61 sediment samples. In the heatmap, * indicates p<0.05, and ** indicates p<0.01.)
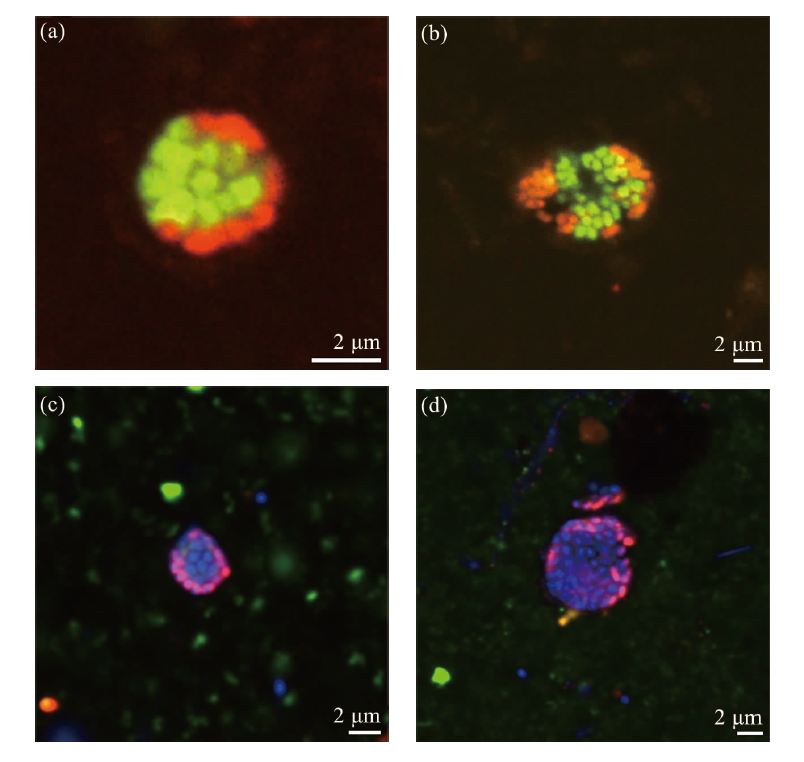
图2 Formosa冷泉样本中ANME-SRB菌团的HCR-FISH图像 (图a显示了由古菌和细菌组成的菌团;图b显示了由ANME-2和SRB组成的ANME-SRB菌团;与DSS探针阳性杂交的ANME-SRB菌团中未观察到与靶向ANME-1(图c)或ANME-3(图d)的探针杂交信号。采用寡核苷酸探针DSS658-R和EUB338-R分别靶向SRB类群和细菌,标记为红色;探针ARCH915-H、ANME-1-350-H、ANME-2-538-H和ANME-3-1249-H分别靶向古菌、ANME-1、ANME-2和ANME-3,标记为绿色;DAPI复染用于细胞核染色,显示为蓝色。)
Fig.2 HCR-FISH images of ANME-SRB consortia in the sediment samples of Formosa cold seep (Figure a illustrates microbial consortia composed of archaea and bacteria. Figure b displays ANME-SRB consortia consisting of ANME-2 and sulfate-reducing bacteria (SRB). No hybridization signals were observed for ANME-SRB consortia that were positively hybridized with the DSS probe when probed with ANME-1-targeting (Figure c) or ANME-3-targeting (Figure d) probes. Oligonucleotide probes DSS658-R and EUB338-R were employed to target SRB and bacteria, respectively, and were labeled red. Probes ARCH915-H, ANME-1-350-H, ANME-2-538-H, and ANME-3-1249-H were used to target archaea, ANME-1, ANME-2, and ANME-3, respectively, and were labeled green. DAPI staining was applied for nuclear staining and appeared blue.)
| 层位 | 细胞团数量/个 | ||
|---|---|---|---|
| ANME-1 | ANME-2 | ANME-3 | |
| 0~2 cm | 0 | 86 | 0 |
| 4~6 cm | 0 | 71 | 0 |
| 8~10 cm | 0 | 73 | 0 |
| 12~14 cm | 0 | 82 | 0 |
| 16~18 cm | 0 | 78 | 0 |
| 平均 | 0 | 78 | 0 |
表3 不同深度沉积物中观察到的ANME细胞团数量
Tab.3 The ANME consortia number observed in the sediments at different depths
| 层位 | 细胞团数量/个 | ||
|---|---|---|---|
| ANME-1 | ANME-2 | ANME-3 | |
| 0~2 cm | 0 | 86 | 0 |
| 4~6 cm | 0 | 71 | 0 |
| 8~10 cm | 0 | 73 | 0 |
| 12~14 cm | 0 | 82 | 0 |
| 16~18 cm | 0 | 78 | 0 |
| 平均 | 0 | 78 | 0 |
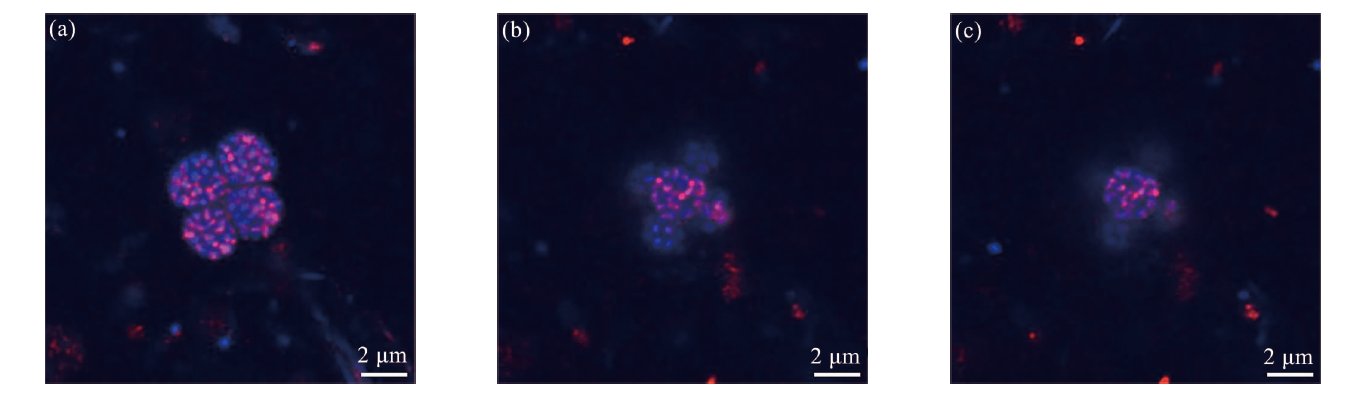
图5 菌团簇的HCR-FISH图像 (菌团与靶向SRB的探针(DSS658-R)阳性杂交,显示为红色; DAPI显示为蓝色。从图a到图c是来自不同z轴的扫描结果。)
Fig.5 The HCR-FISH images of the consortia cluster (The microbial consortia exhibited positive hybridization with the SRB-targeting probe (DSS658-R, red), DAPI staining appeared blue. Figures a to c represent scanning results from different z-axes.)
| [1] |
KNITTEL K, BOETIUS A. Anaerobic oxidation of methane: Progress with an unknown process[J]. Annual Review of Microbiology, 2009, 63: 311-334.
DOI PMID |
| [2] |
PAULL C K, HECKER B, COMMEAU R, et al. Biological communities at the Florida escarpment resemble hydrothermal vent taxa[J]. Science, 1984, 226(4677): 965-967.
PMID |
| [3] | FENG D, QIU J W, HU Y, et al. Cold seep systems in the South China Sea: An overview[J]. Journal of Asian Earth Sciences, 2018, 168: 3-16. |
| [4] | SCHREIBER L, HOLLER T, KNITTEL K, et al. Identi-fication of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade[J]. Environmental Microbiology, 2010, 12(8): 2327-2340. |
| [5] | SCHÖNHUBER W, FUCHS B, JURETSCHKO S, et al. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification[J]. Applied and Environ-mental Microbiology, 1997, 63(8): 3268-3273. |
| [6] | VOLPI E V, BRIDGER J M. FISH glossary: An overview of the fluorescence in situ hybridization technique[J]. Bio-Techniques, 2008, 45(4): 385-409. |
| [7] |
AMANN R, FUCHS B M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques[J]. Nature Reviews Microbiology, 2008, 6(5): 339-348.
DOI PMID |
| [8] |
YAMAGUCHI T, KAWAKAMI S, HATAMOTO M, et al. In situ DNA-hybridization chain reaction (HCR): A facilitated in situ HCR system for the detection of environmental microorganisms[J]. Environmental Microbiology, 2015, 17(7): 2532-2541.
DOI PMID |
| [9] |
YAMAGUCHI T, FUCHS B M, AMANN R, et al. Rapid and sensitive identification of marine bacteria by an improved in situ DNA hybridization chain reaction (quickHCR-FISH)[J]. Systematic and Applied Microbiology, 2015, 38(6): 400-405.
DOI PMID |
| [10] | BHATTARAI S, CASSARINI C, LENS P L. Physiology and distribution of archaeal methanotrophs that couple anaerobic oxidation of methane with sulfate reduction[J]. Microbiology and Molecular Biology Reviews, 2019, 83(3): e00074-18. |
| [11] |
KNITTEL K, LÖSEKANN T, BOETIUS A, et al. Diversity and distribution of methanotrophic Archaea at cold seeps[J]. Applied and Environmental Microbiology, 2005, 71(1): 467-479.
PMID |
| [12] | NAUHAUS K, ALBRECHT M, ELVERT M, et al. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate[J]. Environ-mental Microbiology, 2007, 9(1): 187-196. |
| [13] | NIU M Y, FAN X B, ZHUANG G C, et al. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea[J]. FEMS Microbiology Ecology, 2017, 93(9): 105-110. |
| [14] |
RUFF S E, BIDDLE J F, TESKE A P, et al. Global dispersion and local diversification of the methane seep microbiome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(13): 4015-4020.
DOI PMID |
| [15] | 孙瑜, 牛明杨, 刘俏, 等. 南海Formosa冷泉区沉积物微生物多样性与分布规律研究[J]. 微生物学报, 2022, 62(6):2001-2020. |
| SUN Y, NIU M Y, LIU Q, et al. Diversity and distribution of microorganisms in the sediment of Formosa cold seep in South China Sea[J]. Acta Microbiologica Sinica, 2022, 62(6): 2001-2020. | |
| [16] | JIA Z Y, DONG Y J, XU H, et al. Optimizing the hybridization chain reaction-fluorescence in situ hybridization (HCR-FISH) protocol for detection of microbes in sediments[J]. Marine Life Science & Technology, 2021, 3(4): 529-541. |
| [17] | BOLYEN E, RIDEOUT J R, DILLON M R, et al. Repro-ducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nature Biotechnology, 2019, 37(8): 852-857. |
| [18] |
QUAST C, PRUESSE E, YILMAZ P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools[J]. Nucleic Acids Research, 2013, 41(Database issue): 590-596.
DOI PMID |
| [19] | WEI T, SIMKO V, LEVY M, et al. corrplot: Visualization of a correlation matrix[CP]. (2017-10-16). https://CRAN.R-project.org/package=corrplot. |
| [20] | KOLDE R. pheatmap: Pretty Heatmaps[CP]. (2022-10-14). https://CRAN.R-project.org/package=pheatmap. |
| [21] |
CHOI H M T, CHANG J Y, TRINH L A, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression[J]. Nature Biotechnology, 2010, 28(11): 1208-1212.
DOI PMID |
| [22] | MATSUBAYASHI M, SHIMADA Y, LI Y Y, et al. Phylogenetic diversity and in situ detection of eukaryotes in anaerobic sludge digesters[J]. PLoS One, 2017, 12(3): e0172888. |
| [23] |
MOTER A, GÖBEL U B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms[J]. Journal of Microbiological Methods, 2000, 41(2): 85-112.
DOI PMID |
| [24] | LEVSKY J M, SINGER R H. Fluorescence in situ hybridization: Past, present and future[J]. Journal of Cell Science, 2003, 116(14): 2833-2838. |
| [25] | BOETIUS A, RAVENSCHLAG K, SCHUBERT C J, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane[J]. Nature, 2000, 407(6804): 623-626. |
| [26] |
PERNTHALER A, DEKAS A E, BROWN C T, et al. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(19): 7052-7057.
DOI PMID |
| [27] | LASO-PÉREZ R, WU F B, CRÉMIÈRE A, et al. Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome[J]. Nature Microbiology, 2023, 8(2): 231-245. |
| [28] |
MAIGNIEN L, PARKES R J, CRAGG B, et al. Anaerobic oxidation of methane in hypersaline cold seep sediments[J]. FEMS Microbiology Ecology, 2013, 83(1): 214-231.
DOI PMID |
| [29] |
STOKKE R, ROALKVAM I, LANZEN A, et al. Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments[J]. Environmental Microbiology, 2012, 14(5): 1333-1346.
DOI PMID |
| [30] |
KEVORKIAN R T, CALLAHAN S, WINSTEAD R, et al. ANME-1 archaea may drive methane accumulation and removal in estuarine sediments[J]. Environmental Microbiology Reports, 2021, 13(2): 185-194.
DOI PMID |
| [31] |
RISTOVA P P, WENZHÖFER F, RAMETTE A, et al. Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea)[J]. ISME Journal, 2015, 9(6): 1306-1318.
DOI PMID |
| [32] | CHEN Y, XU C L, WU N Y, et al. Diversity of anaerobic methane oxidizers in the cold seep sediments of the Okinawa trough[J]. Frontiers in Microbiology, 2022, 13: 819187. |
| [33] | MCGLYNN S E, CHADWICK G L, KEMPES C P, et al. Single cell activity reveals direct electron transfer in methanotrophic consortia[J]. Nature, 2015, 526(7574): 531-535. |
| [34] |
WANKEL S D, ADAMS M M, JOHNSTON D T, et al. Anaerobic methane oxidation in metalliferous hydrothermal sediments: Influence on carbon flux and decoupling from sulfate reduction[J]. Environmental Microbiology, 2012, 14(10): 2726-2740.
DOI PMID |
| [35] | MCILROY S J, LEU A O, ZHANG X Q, et al. Anaerobic methanotroph ‘Candidatus Methanoperedens nitroreducens’ has a pleomorphic life cycle[J]. Nature Microbiology, 2023, 8(2): 321-331. |
| [36] |
ORPHAN V J, HOUSE C H, HINRICHS K U, et al. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(11): 7663-7668.
DOI PMID |
| [37] |
KRUKENBERG V, RIEDEL D, GRUBER-VODICKA H R, et al. Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia[J]. Environmental Microbiology, 2018, 20(5): 1651-1666.
DOI PMID |
| [38] |
KLEINDIENST S, RAMETTE A, AMANN R, et al. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments[J]. Environmental Microbiology, 2012, 14(10): 2689-2710.
DOI PMID |
| [1] | 何心怡, 刘倩, 李小虎, 李正刚, 王浩, 朱志敏, 李怀明. 深海多金属结核与周围沉积物中的微生物群落结构特征及其功能[J]. 海洋学研究, 2025, 43(1): 34-46. |
| [2] | 王添翼, 董彦辉, 初凤友, 石学法, 李小虎, 苏蓉, 章伟艳. 太平洋深海富稀土沉积物的分类及成因[J]. 海洋学研究, 2024, 42(1): 23-35. |
| [3] | 邬欣然, 董彦辉, 李正刚, 王浩, 章伟艳, 李怀明, 李小虎, 初凤友. 东太平洋CC区深海稀土资源潜力:沉积物地球化学标志[J]. 海洋学研究, 2023, 41(4): 46-56. |
| [4] | 任世军, 张立, 王红光, 张庆红, 庄彤辉, 魏娜, 宋继红, 程露娴, 王悠, 母清林. 绿色石化项目邻近海域表层沉积物PAHs分布及年际变化[J]. 海洋学研究, 2023, 41(3): 83-91. |
| [5] | 陈一宁, 陈鹭真. 滨海蓝碳生态系统的碳库间相互作用研究进展及展望[J]. 海洋学研究, 2023, 41(1): 3-13. |
| [6] | 刘莉萍, 初凤友, 郭磊, 李小虎. 海底天然气水合物及冷泉流体渗漏的原位观测技术[J]. 海洋学研究, 2023, 41(1): 26-44. |
| [7] | 姚华波, 张朝晖, 金海燕, 陈建芳. 长江口和浙江近岸海域表层沉积物中颗粒磷的形态分布和影响因素[J]. 海洋学研究, 2022, 40(4): 73-81. |
| [8] | 林俊川, 孔德明, 陈法锦, 黄超, . 北部湾沉积物记录的近千年以来气候环境变化[J]. 海洋学研究, 2022, 40(3): 49-61. |
| [9] | 张丽, 杨秀梅, 金海燕, , 朱祖浩, 张秋丰, 戴鑫烽, 陈洁. 有机磷酸酯的污染特征、来源和生态风险:以南海北部湾表层沉积物为例[J]. 海洋学研究, 2022, 40(3): 99-108. |
| [10] | 高抒. 长江口与东海陆架泥质沉积动力过程与环境效应:长期数据采集需求[J]. 海洋学研究, 2021, 39(4): 1-10. |
| [11] | 张从伟, 瞿洪宝, 熊元凯, 韩孝辉, 龙根元, 仝长亮, . 三亚近岸海域表层沉积物微量元素地球化学特征[J]. 海洋学研究, 2021, 39(3): 72-83. |
| [12] | 王苑如, 崔鸿鹏, 李继东, 孙栋, 王春生, 杨娟. 西太平洋多金属结核区表层沉积物细菌群落结构及其对沉积扰动的响应[J]. 海洋学研究, 2021, 39(2): 21-32. |
| [13] | 单红仙, 魏志明, 张民生, 贾永刚. 探头尺寸对FFP确定海底沉积物性质影响的原位试验研究[J]. 海洋学研究, 2020, 38(3): 83-91. |
| [14] | 陆怡, 初凤友, 董彦辉, 朱志敏, 朱继浩, 鲁江姑. 南海东北部陆坡结核成因及对冷泉活动的指示[J]. 海洋学研究, 2020, 38(2): 16-25. |
| [15] | 孟凡盛, 倪建宇, 姚旭莹. 西太平洋马尔库斯-威克海山区沉积物中生物硅含量分布[J]. 海洋学研究, 2019, 37(4): 60-67. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||